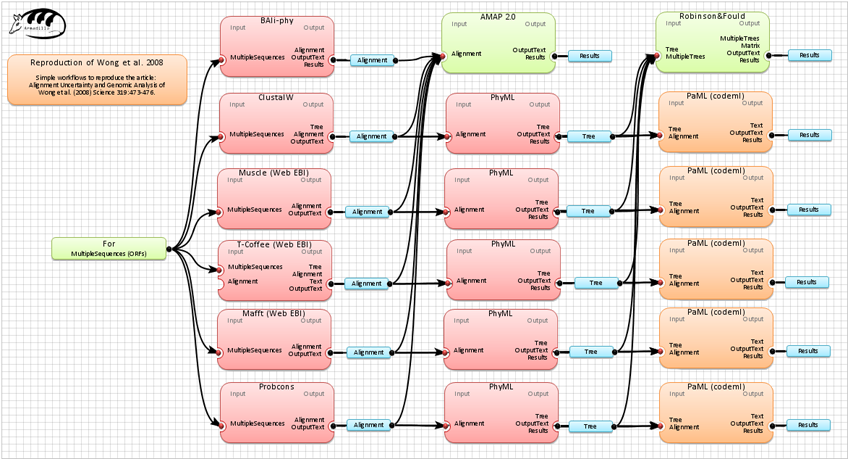
A sample phylogenetic workflow reproducing the work of Wong et al.(2008). Science 319:473-476.
Authors
Etienne Lord*, Mickael Leclercq, Alix Boc, Abdoulaye B. Diallo and Vladimir MakarenkovDépartement d’informatique, Université du Québec à Montréal, PO. Box 8888, Downtown Station, Montreal, Qc, H3C 3P8.
See the new research paper :
Lord E , Leclercq M , Boc A , Diallo AB , Makarenkov V (2012)
Armadillo 1.1: An Original Workflow Platform for Designing and Conducting Phylogenetic Analysis and Simulations
PLoS ONE 7(1): e29903. doi:10.1371/journal.pone.002990
Armadillo 1.1: An Original Workflow Platform for Designing and Conducting Phylogenetic Analysis and Simulations
PLoS ONE 7(1): e29903. doi:10.1371/journal.pone.002990
Summary
Armadillo is a workflow platform dedicated to phylogenetic as well as general bioinformatics analysis. A number of important computational biology tools have been included in the first release. The Armadillo platform is open-source and allows users to develop their own modules and/or integrate existing computer applications. Different complex bioinformatics tasks can be modeled and presented in a single workflow without any prior knowledge of programming techniques. Thus, Life Science students and researchers can develop themselves, with minimum effort, workflow prototypes in order to automate their routines on local workstations. The following popular bioinformatics tasks can be performed using Armadillo:- Multiple sequence alignment (MSA)
- Evolutionary model inference
- Horizontal gene transfer detection
- Inference of phylogenetic trees using Distance, Maximum Likelihood, Maximum Parsimony and Bayesian methods
- Automatic BLAST queries (using the NCBI server)
- Phylogenetic tree manipulation using the PHYLIP package
- Visualisation of phylogenetic trees and networks
Why use Armadillo?
- Automatic data collection of your data in a SQlite compatible database
- Integrated biological sequence and phylogenetic tree manager
- No programming skills required
- Standard sequence and tree formats supported (FASTA, PHYLIP, Newick)
- Automatic conversion of biological sequence data using ReadSeq
- Integrated biological sequence viewer and phylogenetic tree viewer using PhyloWidget
- What You See Is What You Get!
Not in Armadillo? Report a bug.
Note:Armadillo is a work in progress. As such, we'll try to correct as soon as possible any bugs that have been reported.
armadillo.workflow@gmail.com
Need more processing power or other methods for your phylogenetic reconstruction?
Try T-REX, our web interface on this website.
armadillo.workflow@gmail.com
Need more processing power or other methods for your phylogenetic reconstruction?
Try T-REX, our web interface on this website.
System requirements
| Project name | Armadillo
Workflow System |
| Minimum system requirement | Windows version: Windows XP or better. Apple-Macintosh version: Mac OSX Leopard or Lion. Intel Core2 processor, 512mb of RAM 500 Mb of disk space |
| Last version | November 2011 : v1.1 |
| Embedded database system | SQLite v3.6.2 Xerial SQLite JDBC |
| Supporting organisms |   |
Created with Processing.
